Python
Overview
Python is available on the cluster. Depending on the users experience and needs, different distributions are available. Below is an non-exhaustive list of installed distributions:
- Anaconda2
- Anaconda3
- Python
- Biopython
Information, such as available versions, on these distributions can be had using the command
where one has to replace the distribution-name with the name of the distribution on is interested in. For example, when being interested in a specific version of the Python distribution.
will list all the Python versions of the Python distribution we have installed. To activate the version, one needs to enquire about the details of a specific version, load the prerequisites (e.g. compiler, MPI library, etc.) and the Python distribution. E.g. when being interested in Python version 3.6.6 the command
will provide information for which combination of compiler, CUDA and/or MPI library that version of Python is available. Note that the latest Pythonmodules lack packages such as MPI4PY, numpy and scipy. For this you need to load a SciPy-bundle. See below for details.
Please note to utilise a distribution that requires CUDA, you need to have access to an LU Local or LVIS (HPC Desktop On-Demand) project. For more information on how to use the module system consult our separate guide on using the installed software.
Recommended use
Depending on the needs of the user, we recommend different distributions.
Anaconda distributions
For users without special requirements, the recommended distribution is the Anaconda. Anaconda is provided for Python versions 2 and 3. The modules are named Anaconda2 resp. Anaconda3. These are also the recommended distribution for users who whish to use the interactive environment spyder, though spyder is also provided for a few selected versions of the Python distribution.
To get the full functionality of the Anaconda environment should use the following commands:
This enables the full functionality of the conda command.
Creating conda environments
To be able to create reproducable and custom environments with specific versions of Python, Numpy or any other packages. It is encouraged to create a specific conda environment for this. To create a new conda environment make sure that you have activated the conda command with
Now you can create a new environment with the following command:
This will start the process of creating a new environment:
Collecting package metadata (current_repodata.json): done
Solving environment: done
## Package Plan ##
environment location: /home/bmjl/.conda/envs/myenv
Proceed ([y]/n)?
Press y to start the process. When the environment has been created the following is shown:
Preparing transaction: done
Verifying transaction: done
Executing transaction: done
#
# To activate this environment, use
#
# $ conda activate myenv
#
# To deactivate an active environment, use
#
# $ conda deactivate
Activate the new environment using
When activated all package installations, both conda and pip, goes into this environment and will not interfere with the base conda environment.
Python distributions
Users who are planning to do larger scale calculations potentially using MPI4PY or requiring specific compilers and libraries, the recommended distribution is named Python. Please note that this these require loading of pre-requisites, such as compilers, MPI and/or CUDA libraries, as detailed in the Overview section.
Please note that there are several versions of the toolchains such as the foss or intel toolchain. Each with multiple versions of Python. Please choose the one appropriate for you. Note that after having chosen your toolchain use module avail Python to see versions available.
For more information on available toolchains see compiling-code-and-using-toolchains
Python distributions requiring a GCCcore module
Starting in 2019, the pre-requisites and site-package contents of the distributions named Python have changed significantly. The modules can be loaded after loading a GCCcore compiler, which is show in the output of the module spider command. These Python distributions do not contain site-packages depending on an MPI or BLAS library, such as MPI4PY, numpy or scipy. To access such MPI and/or BLAS dependent packages loading a SciPy-bundle-module is required, which require a compiler and an MPI module. Use module spider SciPy-bundle to see what is available.
By default the SciPy-bundle will load a Python 3 module. If you require a Python 2 module, load that prior to loading SciPy-bundle.
Biopython
For users working in bioinformatics on of the Biopython packages is most likely the desired one.
Python site-packages (aka. Python packages)
The distribuitons provided by LUNARC have large number of site-packages (aka. Python packages) installed. Users are encouraged to list the set of installed packages to verify that all needed packages are in place.
Anaconda users can check the installed packages with:
All other installations are using the pip framework:
LUNARC installed packages
LUNARC staff will install packages in the various distributions (mainly in the non-anaconda based ones). If you have a packages that may be of use for other users, or if you are having trouble installing a package, please send a support request for the installation of the package and LUNARC staff will consider it for inclusion in the distribution.
User installed packages
Users are also able to install their own packages by creating their own local repository.
Anaconda Python distributions
In Anaconda (available as a loadable module) the users are free to create their own environment. The user cen either use the pre-installed environment or create an empty environment and populate it with packages that the user chooses using conda create and conda install. Please review: Manage environments (new window) for information on how to use environments in conda.
Please note: The .conda directory where the installed-files are located may become large which may become a problem as the default location for this directory is in /home/< username >. If space is a problem, users should consider using /lunarc/nobackup instead.
Python installations
In all other installations (default or loadable module) users can install their own packages using pip
This will install the package package_name in the users home-directory. Make sure the installation location of your packages gets added to your PYTHONPATH environment variable (e.g. private module file, sourceing a script, ...). For this to work you need a Python package with a recent version of pip. The use of the --install-option to redirect the installation location is no longer recommended.
Again, please check using pip list or `conda list if the package is already installed in the selected distribution.
Jupyter Lab
LUNARC provides means to run Jupyter Lab in compute nodes through Anaconda.
The following instructions work best using the LUNARC HPC Desktop.
Once you started an interactive session, load Anaconda and start jupyter-lab --ip=$HOSTNAME.
After a few seconds, Jupyter lab outputs logs on the terminal such as the example below:
[I 11:49:07.211 LabApp] JupyterLab extension loaded from /sw/easybuild/software/Anaconda3/2020.11/lib/python3.8/site-packages/jupyterlab
[I 11:49:07.211 LabApp] JupyterLab application directory is /sw/easybuild/software/Anaconda3/2020.11/share/jupyter/lab
[I 11:49:07.214 LabApp] Serving notebooks from local directory: /home/username
[I 11:49:07.214 LabApp] Jupyter Notebook 6.1.4 is running at:
[I 11:49:07.215 LabApp] http://au001:8888/?token=a59a11155605a8038b5d61cdab7fde987d56d244b9284fbe
[I 11:49:07.215 LabApp] or http://127.0.0.1:8888/?token=a59a11155605a8038b5d61cdab7fde987d56d244b9284fbe
[I 11:49:07.215 LabApp] Use Control-C to stop this server and shut down all kernels (twice to skip confirmation).
[W 11:49:07.221 LabApp] No web browser found: could not locate runnable browser.
[C 11:49:07.222 LabApp]
To access the notebook, open this file in a browser:
file:///home/username/.local/share/jupyter/runtime/nbserver-22442-open.html
Or copy and paste one of these URLs:
http://au001:8888/?token=a59a11155605a8038b5d61cdab7fde987d56d244b9284fbe
or http://127.0.0.1:8888/?token=a59a11155605a8038b5d61cdab7fde987d56d244b9284fbe
Notice the line http://au001:8888/?token=a59a11155605a8038b5d61cdab7fde987d56d244b9284fbe must appear in output.
The hostname (here au001) depends on the compute node allocated for the interactive session and the token (a59a11155605a8038b5d61cdab7fde987d56d244b9284fbe) is unique to the jupyter-lab sessions being run.
Note that you cannot use the link with hostname '127.0.0.1' or 'localhost'.
You can open the Jupyter Lab web interface by copying the whole line and pasting in a browser running in the LUNARC HPC Desktop.
Note that you can share the session with any other LUNARC user logged in the LUNARC HPC Desktop by sharing the link with them.
Any user with whom you share this link can read and write all files in any subdirectory from where the Jupyter lab was started. Share the link with caution.
Virtual environments
If the python software environment in Jupyter Lab lacks additional software of python packages, a customized Jupyter Lab python kernel can be a solution.
Building and installing a custom Jupyter Lab kernel
To build a customized kernel, create a new virtual Anaconda environment and install anaconda packages ipykernel and ipython_genutils as well as other anaconda or pip packages required by your application. Finally, activate the Anaconda virtual environment and register the new kernel to Jupyter Lab:
$KERNEL_NAME is the name you choose for the environment (e.g. "tensorflow") and $KERNEL_DISPLAY_NAME is its human-readable name, e.g., "Python (TensorFlow)".
The new Jupyter Lab kernel is now ready to use. Proceed with starting Jupyter Lab as described above, but make sure to activate the conda virtual environment after the interactive session started and before starting Jupyter Lab.
Once installed, the new custom kernel can be selected either on Jupyter Lab web page startup:
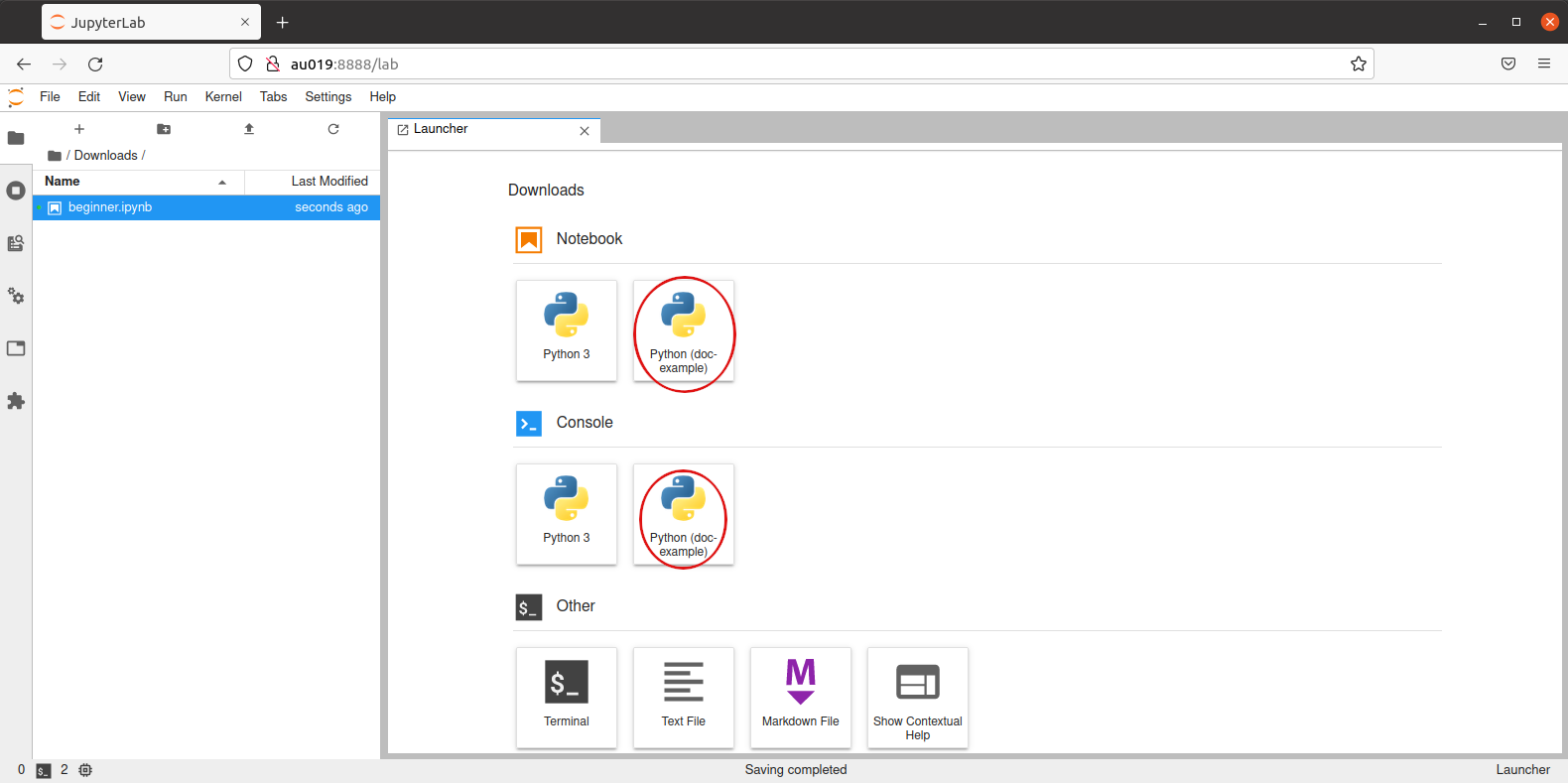
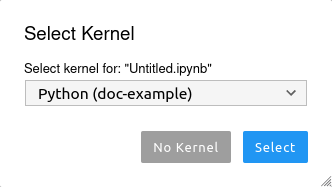
via main menu Kernel/Change Kernel... or on Jupyter Lab startup:
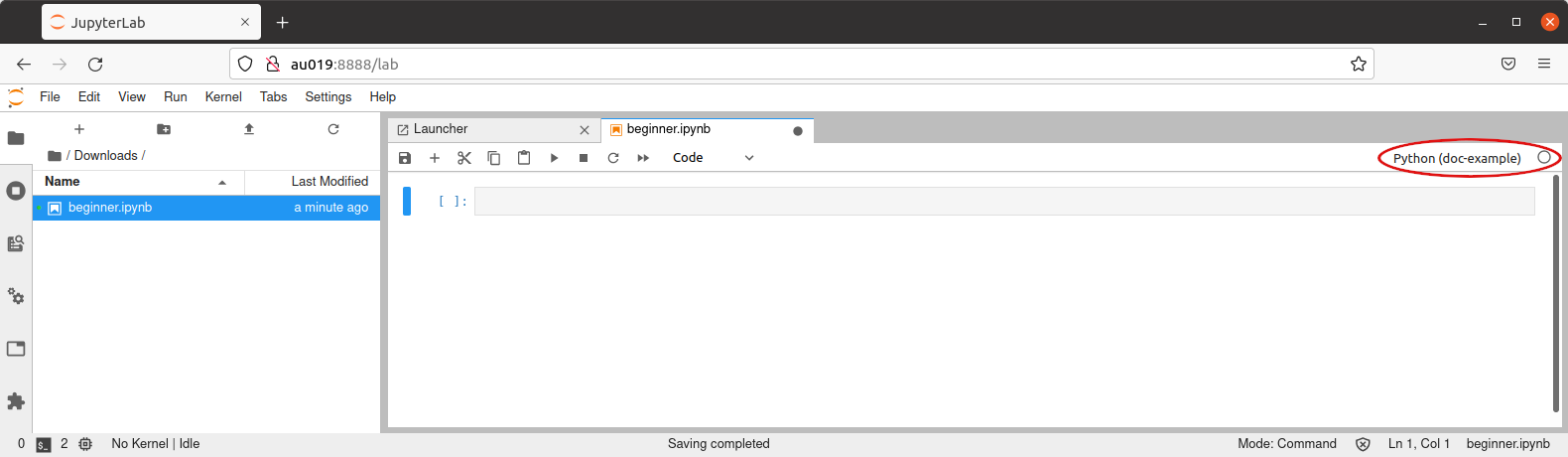
In any case, the kernel in use at any given time is shown on the upper-right corner:
Removing a custom kernel
To remove a custom kernel, activate the corresponding conda environment, and run the command
where $KERNEL_NAME is the name of the kernel to remove.
Author: (LUNARC)
Last Updated: 2022-10-05